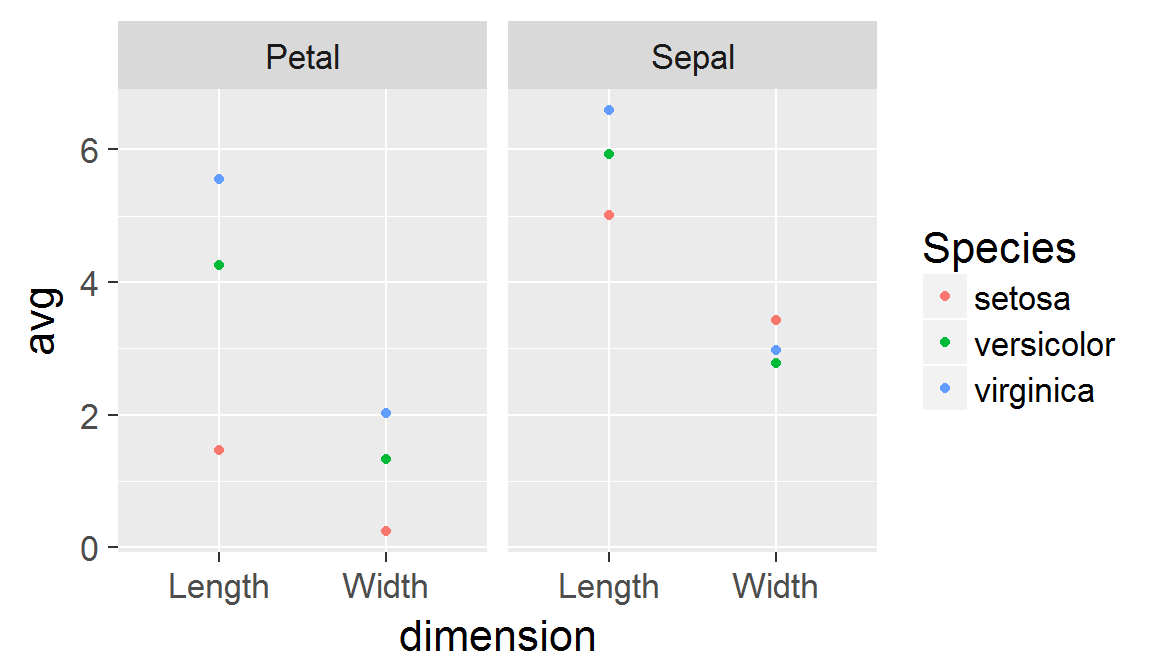
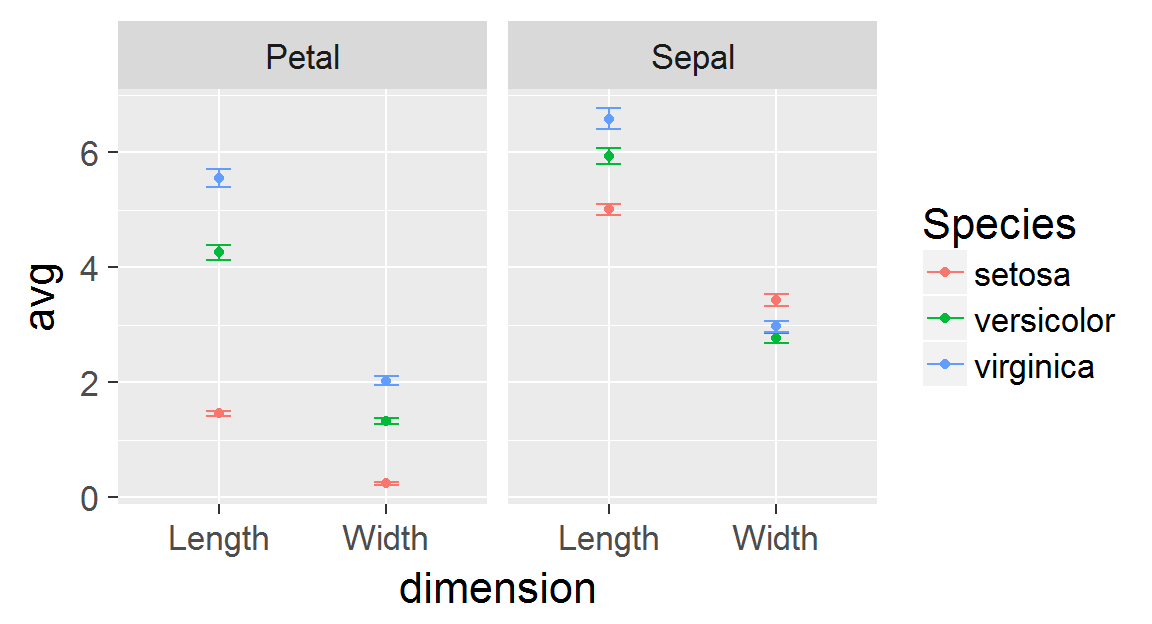
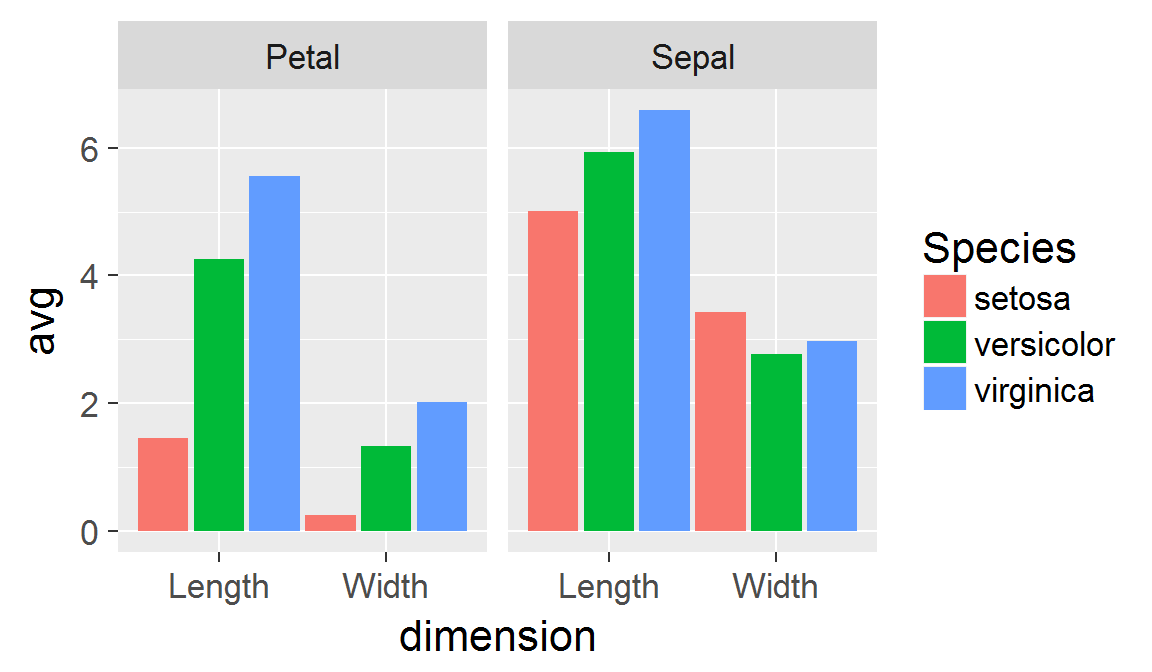
We're going to continue the tour of the ggplot2 package we began last week
We'll explore some more "advanced" features of the package, look at data management issues in the context of ggplot-ing, and make some common types of visualizations.
NB: This lab was made using ggplot2 version 2.2.1, and a few things I'll be using are specific to ggplot2 version >= 2.0. So, if you have an older version installed, you'll need to upgrade to follow along. You can check your version using the command packageVersion("ggplot2")
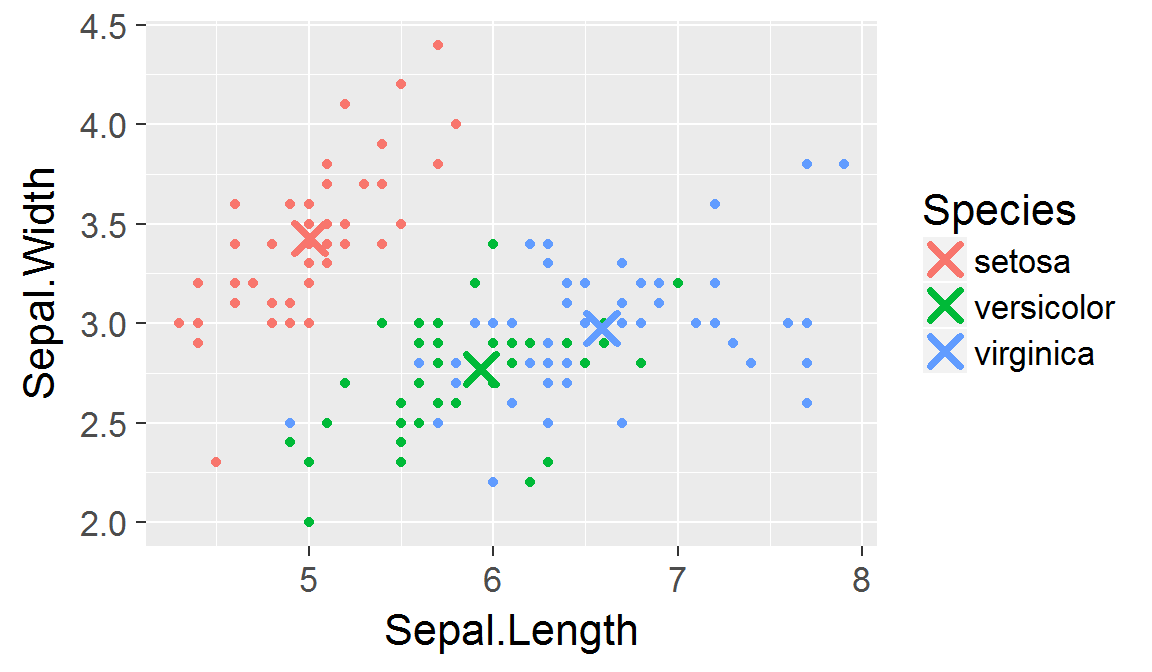
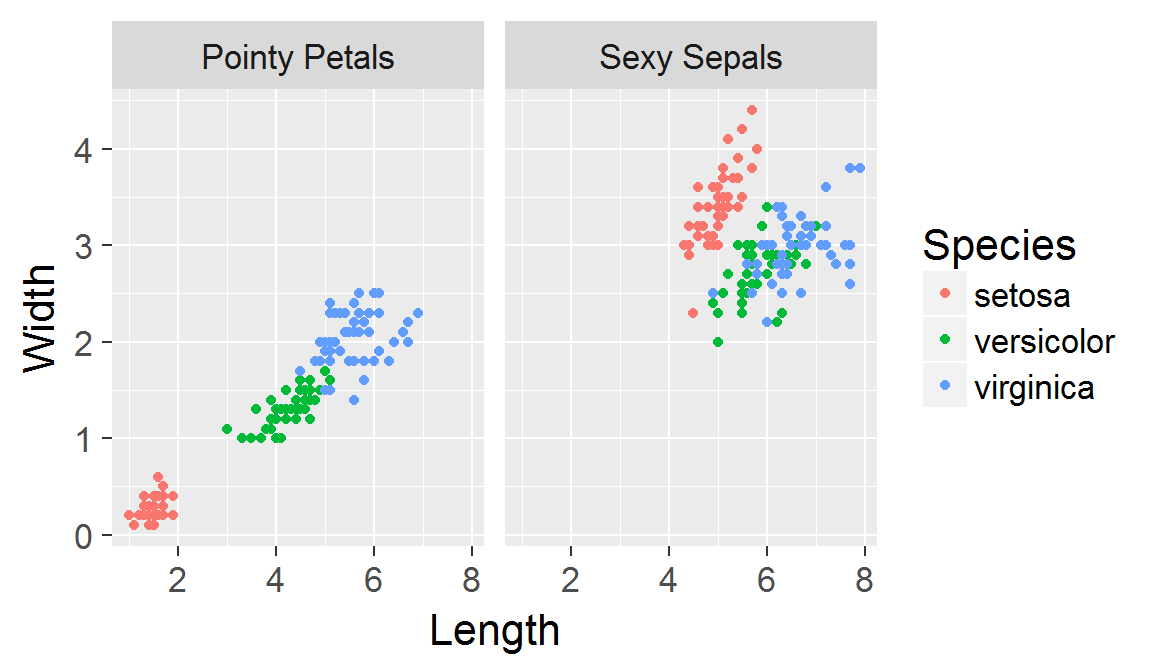
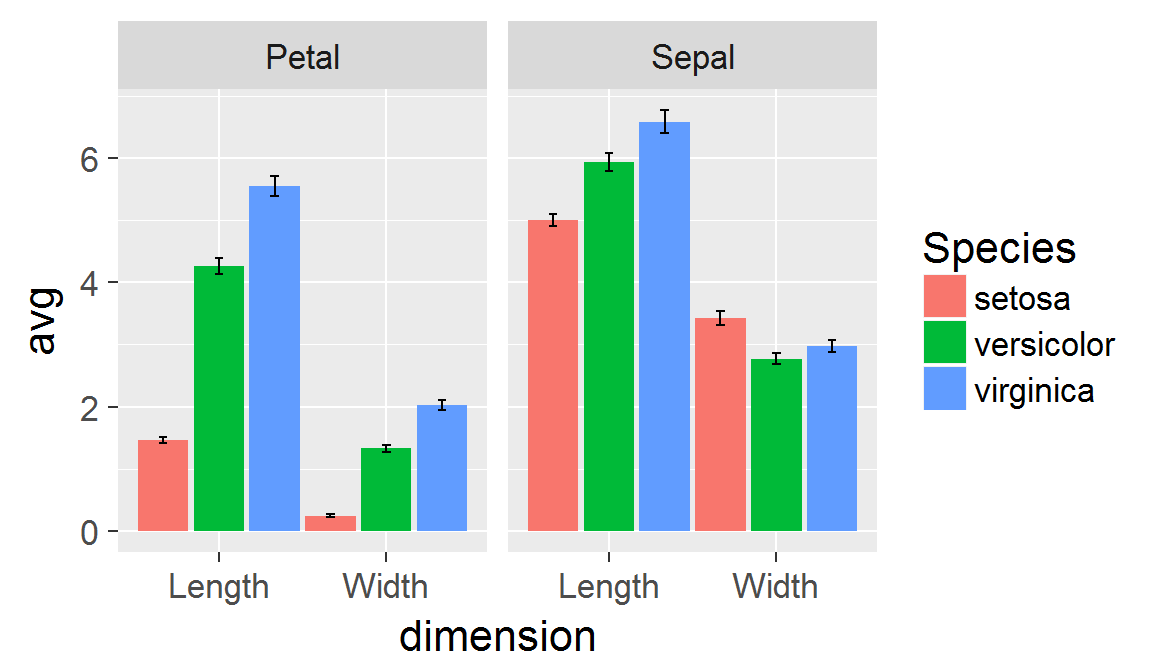
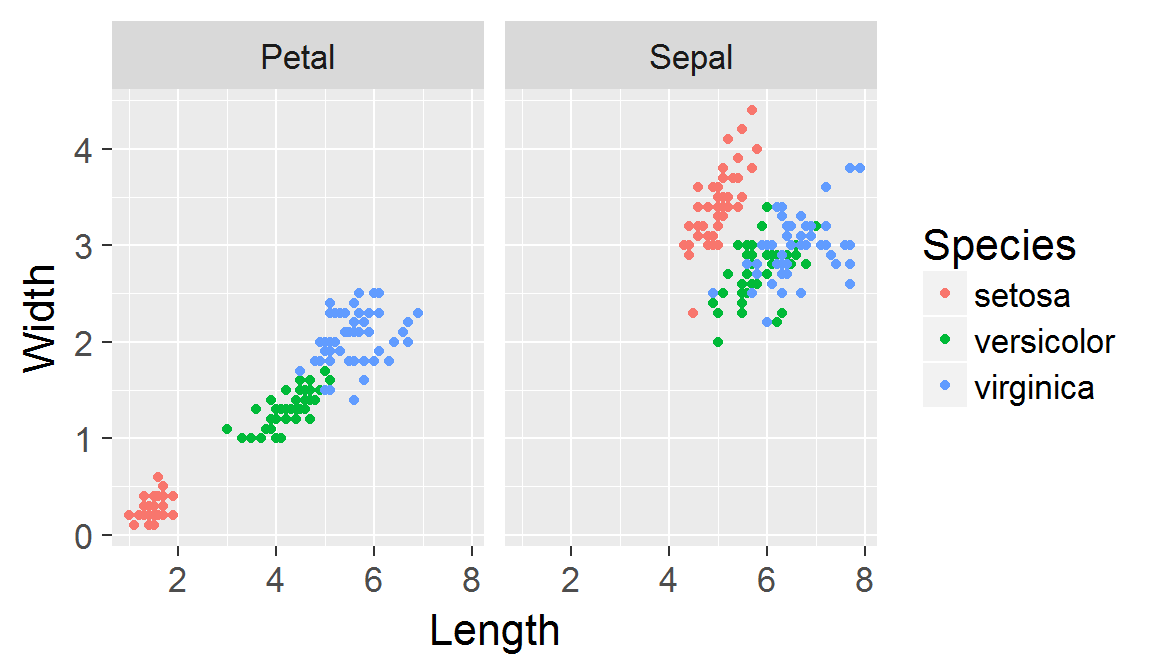 Panels created with faceting share the same scales!
Panels created with faceting share the same scales!